- Name
- Amanda Zrebiec
- Amanda.Zrebiec@jhuapl.edu
- Office phone
- 240-592-2794
An effort by scientists from across Johns Hopkins University and Health System to sequence the genome of SARS-CoV-2, the virus causing COVID-19, has produced a study characterizing the early introduction of the virus into the Washington, D.C., metropolitan area.
Among several findings, the study points to multiple entries of the virus into the region but similar clinical presentation, an indication that, despite small genetic differences, circulating viruses are likely causing identical disease. The research also provides information about the progression of cases as quarantine procedures were implemented, and ways to study the efficacy of an eventual vaccine.
Initiated by the work of molecular biologists at the Johns Hopkins Applied Physics Laboratory and using software and approaches developed, in part, at APL, the team's early takeaways are featured in the study, which is awaiting peer review. The study was funded by the JHU COVID-19 Research Response Program, which provided seed grants to rapid response teams to find solutions and increase knowledge amid the COVID-19 pandemic.
The Johns Hopkins Health System diagnosed more than 37% of early COVID-19 cases in the state of Maryland, from patients living across the Baltimore-Washington metro area. Genomes for 114 of those diagnosed positive samples were sequenced, and those patients' disease presentation and outcomes were analyzed.
Through that analysis, the team observed multiple likely introductions of the SARS-CoV-2 virus into the region, as well as in many patients with no travel or sick contact history, an indication that community transmission was well established in the area in early March. What they did not detect was a distinct link between viral genetic variations and patient outcomes, such as hospital or ICU admission—meaning they found no likely connection between the virus' genealogy and the severity of infection in patients.
"Despite observing genetic diversity in samples from the region, it appears to be clinically the same coronavirus," said Peter Thielen, a molecular biologist in APL's Research and Exploratory Development Department and one of the co-lead authors on the study with Shirlee Wohl, a postdoctoral fellow in the Department of Epidemiology at the Johns Hopkins Bloomberg School of Public Health. "Our data does not show a linkage between patient outcome and virus genotype."
That connection, consisting of the clinical and genome sequence data, allows the Hopkins team to further disabuse the notion that there are multiple "strains" of the novel coronavirus, some of which are deadlier than others.
The final total of sequenced samples included 40 from hospitalized patients and 22 from patients admitted to the ICU. In short, the authors write, "The diversity of virus genetics, clinical symptoms, and patient outcomes suggests that viral mutations are not the main driver of clinical presentation," and they "observed that severe cases, defined as admission to the ICU (including patients requiring ventilator support), had viral genomes belonging to all five major phylogenetic [groupings]."
"Efforts to control local spread of the virus were likely confounded by the number of introductions into the region early in the epidemic and interconnectedness of the region as a whole," the authors write.
The team also observed that countermeasures for the initial SARS-CoV-2 genome likely apply to all COVID-19 cases.
"Designing vaccines and therapeutics to a single strain, even one with the type of genetic diversity we've observed here, is much more straightforward than a virus that is changing quickly, such as influenza," Thielen explained. "What we observe in the local and global data provides excellent reassurance that research in those areas is on the right track."

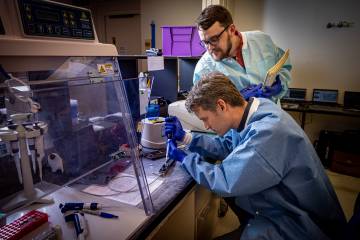
Image caption: Tom Mehoke, a molecular biologist from Johns Hopkins APL, studies sequencing data at the molecular diagnostics laboratory at Johns Hopkins Hospital in late March.
Image credit: Ed Whitman / Johns Hopkins APL
In its paper, the team traces the path of the virus' initial introduction into the region, revealing clinical data from patients with recent travel history to England and elsewhere in North America (including California, Colorado, New York, and Idaho). The researchers also compared the region's genetic diversity to that of others in the U.S. and around the world. The diversity of the viral population described in New York City appears to be similar to that of the Washington, D.C., area, as outbreaks in both regions were seeded multiple times.
Genetic characterization of viral pathogens requires expertise ranging from molecular diagnostics to DNA sequencing technologies, as well as computational biology and epidemiology. The paper's authors include APL researchers Thielen, Tom Mehoke, Jared Evans, Craig Howser, Brian Merritt, Tina Zudock, Kianna Blount, and Amanda Ernlund. Researchers from the Johns Hopkins School of Medicine, Whiting School of Engineering, and Bloomberg School of Public Health were led by Winston Timp, Heba Mostafa, and Stuart Ray.
"This interdisciplinary team represents many specific strengths of Johns Hopkins and is an important contribution to our understanding of the early stage of the COVID-19 pandemic in the U.S.," Thielen said. "As the pandemic continues and countermeasures become available, it will be important to track these genetic changes and identify correlations between virus genetics, clinical presentation, and the potential impact on vaccine efficacy."
The team will continue using viral sequencing for COVID-19 surveillance in the region, comparing samples collected during Maryland's stay-at-home order to those gathered during the state's gradual reopening. That analysis will allow the researchers to better understand the effectiveness of the lockdown and primary modes of viral transmission within the region.