- Name
- Vanessa Wasta
- wasta@jhmi.edu
- Office phone
- 410-955-8236
Johns Hopkins scientists have developed a method involving artificial intelligence to visualize and track changes in the strength of synapses—the connection points through which nerve cells in the brain communicate—in live animals. The technique, described in Nature Methods, should lead to a better understanding of how such connections in human brains change with learning, aging, injury, and disease, the scientists say.
"If you want to learn more about how an orchestra plays, you have to watch individual players over time, and this new method does that for synapses in the brains of living animals," says Dwight Bergles, professor in the Department of Neuroscience at the Johns Hopkins University School of Medicine.
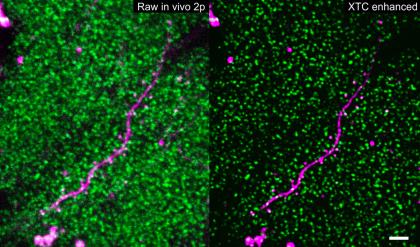
Image caption: Thousands of SEP-GluA2 tagged synapses (shown in green) surround a sparsely labeled dendrite (show in magenta) before and after XTC image resolution enhancement. Scale bar is 5 microns.
Image credit: Xu, Y.K.T., Graves, A.R., Coste, G.I. et al. Nat Methods
Bergles co-authored the study with colleagues Adam Charles and Jeremias Sulam, both assistant professors in the Department of Biomedical Engineering, and Richard Huganir, Bloomberg Distinguished Professor at JHU and director of the neuroscience department. All four researchers are members of Johns Hopkins' Kavli Neuroscience Discovery Institute.
Nerve cells transfer information from one cell to another by exchanging chemical messages at synapses, or junctions. In the brain, the authors explain, different life experiences, such as exposure to new environments and learning skills, are thought to induce changes at synapses, strengthening or weakening these connections to allow learning and memory. Understanding how these minute changes occur across the trillions of synapses in our brains is a daunting challenge, but it is central to uncovering how the brain works when healthy and how it is altered by disease.
To determine which synapses change during a particular life event, scientists have long sought better ways to visualize the shifting chemistry of synaptic messaging, necessitated by the high density of synapses in the brain and their small size—traits that make them extremely hard to visualize even with new state-of-the-art microscopes.
"We needed to go from challenging, blurry, noisy imaging data to extract the signal portions we need to see," Charles says.
To do so, Bergles, Sulam, Charles, Huganir, and their colleagues turned to machine learning, a computational framework that allows flexible development of automatic data processing tools. Machine learning has been successfully applied to many domains across biomedical imaging, and in this case, the scientists leveraged the approach to enhance the quality of images composed of thousands of synapses. Although it can be a powerful tool for automated detection, greatly surpassing human speeds, the system must first be "trained," teaching the algorithm what high quality images of synapses should look like.
In these experiments, the researchers worked with genetically altered mice in which glutamate receptors—the chemical sensors at synapses—glowed green, or fluoresced, when exposed to light. Because each receptor emits the same amount of light, the amount of fluorescence generated by a synapse in these mice is an indication of the number of synapses, and therefore its strength.
As expected, imaging in the intact brain produced low quality pictures in which individual clusters of glutamate receptors at synapses were difficult to see clearly, let alone to be individually detected and tracked over time. To convert these into higher quality images, the scientists trained a machine learning algorithm with images taken of brain slices (ex vivo) derived from the same type of genetically altered mice. Because these images weren't from living animals, it was possible to produce much higher quality images using a different microscopy technique, as well as low quality images—similar to those taken in live animals—of the same views.
This cross-modality data collection framework enabled the team to develop an enhancement algorithm that can produce higher resolution images from low quality ones, similar to the images collected from living mice. In this way, data collected from the intact brain can be significantly enhanced and able to detect and track individual synapses (in the thousands) during multiday experiments.
To follow changes in receptors over time in living mice, the researchers then used microscopy to take repeated images of the same synapses in mice over several weeks. After capturing baseline images, the team placed the animals in a chamber with new sights, smells, and tactile stimulation for a single five-minute period. They then imaged the same area of the brain every other day to see if and how the new stimuli had affected the number of glutamate receptors at synapses.
Although the focus of the work was on developing a set of methods to analyze synapse level changes in many different contexts, the researchers found that this simple change in environment caused a spectrum of alterations in fluorescence across synapses in the cerebral cortex, indicating connections where the strength increased and others where it decreased, with a bias toward strengthening in animals exposed to the novel environment.
The studies were enabled through close collaboration among scientists with distinct expertise, ranging from molecular biology to artificial intelligence, who don't normally work closely together. The researchers are now using this machine learning approach to study synaptic changes in animal models of Alzheimer's disease, and they believe the method could shed new light on synaptic changes that occur in other disease and injury contexts.
"We are really excited to see how and where the rest of the scientific community will take this," Sulam says.
The experiments in this study were conducted by Yu Kang Xu, a PhD student and Kavli Neuroscience Discovery Institute fellow at JHU; Austin Graves, assistant research professor in biomedical engineering at JHU; and Gabrielle Coste, a neuroscience PhD student at JHU. This research was funded by the National Institutes of Health (RO1 RF1MH121539).
Posted in Health, Science+Technology
Tagged neuroscience, molecular biology, biomedical engineering, artificial intelligence







