- Name
- Roberto Molar Candanosa
- rmolarc1@jh.edu
- Office phone
- 443-997-0258
- Cell phone
- 443-938-1944
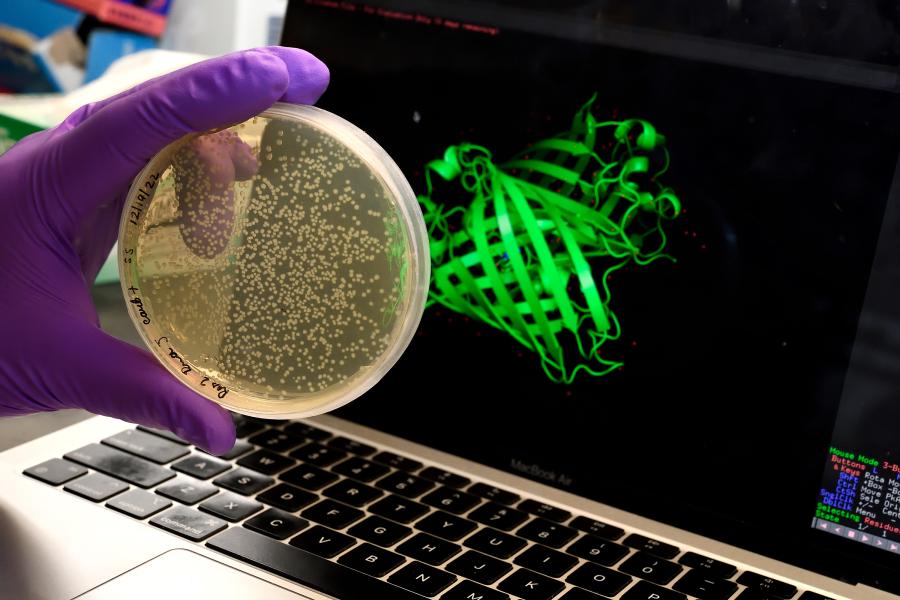
Researchers have discovered a surprising anomaly in the behavior of how proteins form, upending long-held assumptions about the way cells produce these crucial molecules and potentially leading to a better understanding of aging and neurodegenerative diseases in humans.
Contradicting conventional wisdom that proteins can reassemble themselves, Johns Hopkins University biochemists found a significant number of the proteins in E. coli could not, even when the team tried to spark the repairs in the lab with helper proteins called "chaperones."
The finding was stunning, said senior author Stephen Fried, an assistant professor in the Department of Chemistry in the Krieger School of Arts and Sciences who led the research published recently in Proceedings of the National Academy of Sciences.
"The most surprising finding is that there are certain proteins even chaperones can't help," Fried said. "If proteins misfold, we are taught that chaperones are supposed to be able to fix them. But some proteins are like Humpty Dumpties: Once they fall, all the cells' men and horses can't put them back together again."
Proteins are long chains of molecules consisting of smaller components called amino acids. All cells—human or not—contain proteins that execute an endless number of functions including fighting viruses, building tissue, running organs, and producing other types of molecules.
A protein's shape determines its ability to function properly. The way their amino acid chains "fold" or organize into specific three-dimensional structures determines the functions they perform. If unfolded proteins were a necklace of pearls, functional proteins would look like the pearls organized into balls, tubes, and other structures of many shapes and forms.
Genetic mutations and other biochemical mishaps within cells can cause proteins to misfold into dysfunctional structures. In humans, mistakes in protein synthesis and folding can kill neurons and cause Alzheimer's, Parkinson's, and other neurodegenerative diseases associated with aging. But the details of how this behavior damages a cell's functions are still unclear.
Fried hopes the findings help illuminate that process.
"From decades of protein folding research, we know a lot about a very small number of very simple proteins because those were the ones that were amenable to the types of experiments biophysicists were good at," Fried said. "We now have these really amazing technologies in the field to analyze tens of thousands of proteins in one sample, but this technology had never really been deployed to look at folding."
With scientists at Pennsylvania State University, Fried's lab is also working to gain a better understanding of why some proteins can't refold. Their findings, published in Nature Chemistry, show some proteins can only fold properly when a cell's ribosome produces them for the first time. The research also shows subtle mutations could be changing how quickly or slowly a cell builds and folds specific proteins.
Fried is also collaborating with Johns Hopkins neuroscientist Michela Gallagher to study how the E. coli protein findings compare to proteins in the brains of aging rats with memory loss and other cognitive impairments. They hope this will provide insight into how protein folding glitches influence brain disease in humans.
Johns Hopkins authors included Philip To, Yingzi Xia, Sea On Lee, Taylor Devlin, and Karen G. Fleming. Penn State researchers included Yang Jiang, Syam Sundar Neti, Ian Sitarik, Priya Pradhan, Yingzi Xia, Squire J. Booker, and Edward P. O'Brien.
The work was supported by NIH Director's New Innovator Award (DP2GM140926), the NSF Division of Molecular and Cellular Biology (MCB2045844). NIGMS (R01GM079440), and NIH training grant (T32GM008403).
Posted in Science+Technology
Tagged biology, neuroscience, chemistry, biochemistry