Audrey Branch is trying to learn more about aging by studying old and young brains. Specifically, she's interested in how cells connect to form memories and what might be going wrong with those connections when older people start to forget things.
Until recently, getting at that question meant months of tedious specimen preparation. And even then, the very prep that made getting a glimpse of the brain's core possible—slicing what's already tiny into thousands of pieces—very likely destroyed the delicate connections the Johns Hopkins neuroscientist needed to see.
That changed this spring when a new, three-dimensional microscope arrived at the university's Homewood campus, a cutting-edge tool that not only condenses what had been months of work into just hours, but allows researchers unprecedented views of organs, tissue, and even live specimens.
Just practicing with it, Branch knew it was a game-changer. She cried when she saw the first pictures of a mouse brain, its individual neurons glowing red, and its spindly dendrites, too—showing quite clearly the links between those cells.
"It feels so amazing to see the brain in a way that no one has ever seen it before," she said. "It's pretty much the greatest thing I've ever experienced in science."
The selective plane florescence light sheet microscope arrived on campus in April, one of the first in operation on the East Coast and the only one in Maryland. Purchased with a grant from the National Institutes of Health, it cost $360,000.
Unlike other microscopes, this one illuminates specimens from the side, shooting two perfectly aligned planes of light across an object, illuminating a wafer-thin slice of the whole while the camera captures the image—thousands of times over as the specimen moves through the light. When the images are displayed together, the result is a three-dimensional image or video clip of the full object, sort of like the more familiar CAT scan.
The technology is very new, but Michael McCaffery, director of the university's Integrated Imaging Center, expects researchers everywhere will be using it within a few years. Just among the Johns Hopkins community, word of the light sheet is already out and scientists have been lining up to use it—even if that requires the minor inconvenience of bringing specimens over from the medical campus.
"People really want to use this," McCaffery said. "It fills a niche that until now was unavailable at Hopkins. Simply, there was no instrument that allowed a researcher to take a whole organ, brain, or cardiac muscle, and image them in three-dimensions, in their entirety."
The light sheet is the latest advance in modern microscopy—a world that's been evolving since fluorescence microscopy became the standard in the 1960s. Now, most researchers use confocal microscopes, which use lasers to illuminate a sample point by point—only extremely tiny samples will work—then create computerized images, pixel by pixel.
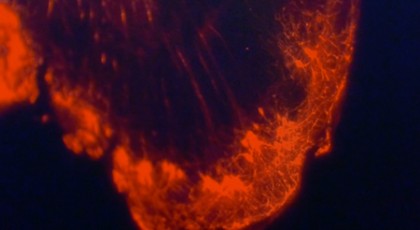
Image caption: A still image captures one side of the story of interconnected cells—but the image comes to life and reveals far greater information when represented in 3-D on a computer screen
Image credit: Office of Communications
Confocals produce vivid, high-resolution images, but the sample size limitations—nothing thicker than about 70 microns, which is about as wide as a strand of human hair—severely handicapped scientists.
The new light sheet allows samples up to 12 to 15 millimeters, or about a half an inch. Researchers can study much larger samples, even entire organs. And because the samples don't have to be cut up, researchers like Branch who are interested in how cells, ducts, or veins connect have a chance to observe them, unspoiled.
"It's a very big deal for researchers, particularly those interested in the science of connectomics," McCaffery said. "Mapping the neuronal connections of the brain is the holy grail of neurology."
It's certainly Branch's holy grail.
Branch is a Kavli Neuroscience Discovery Institute fellow working in the Krieger School of Arts and Sciences. She wants to know how newborn neurons, which are key to making memories, connect to other cells in the brain—and how those connections might change as people age.
Scientists know the number of newborn neurons declines with age, and that likely has something to do with why short-term memory declines with age. What Branch wants to do is audit these newborn cells in a young brain, determining how many there are, where they are, and what other cells they communicate with. She can compare that with an older brain and possibly see which connections have broken when memory loss occurs. If she can target the broken connections, there could be a way to treat the area with a drug and stop or slow cognitive decline.
Branch has been practicing on the light sheet with mouse brains, and she plans to formally investigate her hypothesis with rat brains, which are bigger and more human-like.
If she didn't have the light sheet, Branch would have to slice the brain, which is about the size of an olive pit, into tissue-thin sections—about 250 pieces. Each slice would need to be stained, mounted onto a slide, and then imaged. Each of those images would need to be manually assembled into a composite to approximate the whole.
All of this work would take about a month. Since Branch's experiment involves 30 brains, it would take her about two and a half years, "if," she says, "that's all I did day in and day out."

Image caption: Audrey Branch preps a new sample for study under the microscope; behind her, a computer screen displays images already captured in 3-D
Image credit: Will Kirk / Johns Hopkins University
Worse yet, by slicing the brain, she would lose most of the newborn neurons she needed to find, and probably all of the connections. She figures if she had marked 50 newborn neurons, she'd be lucky to find five.
"It would be impossible to find the connections," she says. "And it would be impossible to get an idea of who each of those cells is talking to. Maybe it's not important, but I'm guessing that's not the case. Neurons in isolation aren't interesting; it's who they're talking to, it's how they're wired.
"I was just going to have to estimate. I'd have missed a lot of the picture, and that's all anyone's been able to do."
Guy Bar-Klein, a neuroscientist working in the Hal Dietz Lab at the School of Medicine, has been crossing town to spend time at Homewood's Dunning Hall with the light sheet to study blood vessels in the heart and brain, hoping to better understand what causes aneurysms.
Without the light-sheet technology, his view would be limited to a minuscule section of tissue, much too small to get a true sense of its vasculature. Now, he has been looking at samples with intact blood vessels, making it possible to spot and track aneurysms—and possibly pinpoint the underlying issues that caused it to form.
"It's very exciting," Bar-Klein said. "I think it gives us a very substantial advantage in understanding the signaling involved in aneurysm formation."
Michael Noë, a pathology resident who studies pancreatic cancer, hopes the light sheet's three-dimensional perspective will allow him to see relationships between tumors and the surrounding nerves and blood vessels. Tumors often grow around nerves, and Noë expects the new perspective of cancerous ducts and nerves could shed light on why.
"For almost 200 years, pathologists looked at tissue the same way," he says. "Three-dimensional is almost a whole new world for us. There is a lot of excitement in the department of pathology to apply this technology for the first time to human samples."
Before researchers can view tissue of any sort with the light-sheet, their samples must be treated to make them translucent, so the microscope's light can pass through and create an image. Noë has developed a protocol for clearing human tissue and tumors, work he's hoping to publish.
Branch expects to have 3-D images of all 30 of her rat brains in three to six months.
She'll see every newborn neuron. She'll see each dendrite. And hopefully, she'll find answers – she already knows she'll find more questions.
"The technology makes it easier to have confidence about our findings," she says, "It also opens up an opportunity to ask even more questions — things that before, we didn't even know we could ask."
Posted in Health, Science+Technology
Tagged biology, neuroscience, microbiology







