Peter Thielen and Thomas Mehoke, molecular biologists at the Johns Hopkins Applied Physics Laboratory, are working to sequence the genomes of different variations of SARS-CoV-2, the virus that causes the illness COVID-19, in order to track its mutations as it spreads. They joined Sarah LaFave, a PhD student at the Johns Hopkins School of Nursing, to discuss how they are carrying out their work and how it can inform the evolving response to the pandemic. The conversation has been edited for length and clarity.
What is genomic sequencing and why is it important to understand the genomic sequence of COVID-19?
Thielen: Genomic sequencing is a technique that allows us to read and interpret genetic information found within DNA or RNA. When we look at virus genome sequences from patient samples that test positive for COVID-19, we're interested in understanding where their version of the virus originated. For example, does the virus look similar to how it looks in Washington State? Or in New York? Or in Europe? Right now, we're working to analyze many genome sequences from SARS-CoV-2, the virus that causes COVID-19, that are circulating in the Baltimore area and in Maryland. Our goal is to understand how the virus is evolving as it spreads. So far, there are over 1,000 COVID-19 genomes that have been published worldwide.
Mehoke: As the outbreak progresses, this work will help us understand how well the virus is contained in Maryland.
So by looking at the genomic sequence of the virus causing COVID-19 in a particular person's sample, you can begin to understand how the virus is spreading because the genomic sequence looks a little different as the virus mutates and spreads in different geographic areas?
Thielen: Right. The initial two COVID-19 sequences that we've analyzed suggest that the viruses circulating locally have small genetic changes that are different from the ones circulating in, for example, Washington State. This suggests that the virus here in Maryland may have been imported from geographic locations other than China, because we expect that cases in Washington State were originally introduced from China. Logically, we're expecting that we would see more virus imported from European countries because of travel patterns. But we'll know more about whether or not that is true as we generate more sequences.
Are there other potential public health uses of your data?
Mehoke: Yes, you could use the genomic sequence to estimate the actual infected population size. So rather than just determining the number of people who have tested positive, from the genomes that we are seeing, we can estimate the total number of positive cases in the state, and that can give us a better understanding up-front of the scope of the problem. This is especially important given the limited scale of testing in some locations.
It seems like speed must be a really important part of this work. How was your lab able to start this research so quickly?
Thielen: For the last several years, our center and our partners have been developing capacity to analyze flu in preparation for whatever the next outbreak might be. We've had an eye on different pathogens that could lead to pandemics, and we can quickly modify all our resources.

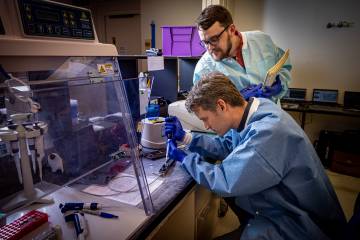
Image caption: APL biologist Tom Mehoke reviews the DNA sequencing analysis of SARS-CoV-2, the virus causing COVID-19, at the molecular diagnostics laboratory at Johns Hopkins Hospital.
Image credit: Johns Hopkins APL / Ed Whitman
Mehoke: We don't want to be writing papers about this a year from now saying, "This is how the virus might be spreading." We want to get data into the hands of people who can do something with it right now. We're trying to make it as quick and as automated as possible to go from swabbing someone's nose to getting a genomic sequence.
How do you acquire the genetic material and then run the genomic sequence?
Mehoke: First, someone at the hospital swabs a patient's nose, and we pull genomic material out of that sample. We do this work on a little handheld DNA sequencer, which is smaller than a phone, that connects over USB to a laptop.
Thielen: We use data analysis software on our laptops to sequence the genome, and then upload the sequences directly into international databases. The databases can be accessed by people in the viral genomics community so that scientists around the world can have access to the widest array of data as possible.
How do the genomic sequencing collaborations between scientists across countries work?
Thielen: There is an incredible amount of international collaboration. We're working with the NIH's Fogarty International Center to collaborate with researchers in low- and middle-income countries to perform this type of work in those settings. Low- and middle-income countries are usually "data dark spots" during an outbreak. Now that these new technologies are available, low- and middle-income countries can pretty easily set up sequencing capacity using just a laptop computer and a handheld sequencer, without the need for large research laboratories. We are currently working to get our international collaborators up and running with the same capacity that we have at Johns Hopkins.
Mehoke: We're also working closely with the international ARTIC network. Their philosophy is that scientists should share data as much as possible and get information out quickly. We're very supportive of that philosophy. We're having conversations within and outside of Hopkins to share knowledge as much as we can.
Posted in Health, Science+Technology, Voices+Opinion
Tagged genomics, q+a, coronavirus