Mark Foster is an applied physicist by training, one adept at building advanced optical systems that are capable of imaging, for instance, a single neuron firing inside a mouse's brain. Standing at the whiteboard of his Homewood campus office, he is reminded that he is not, however, a skilled artist.
"Well, that doesn't look much like a tree," says the associate professor of electrical and computer engineering, drawing a green squiggle in dry erase marker. "Ehh, I can draw better," he continues, erasing and trying again. Foster is playing this pseudo-game of Pictionary to explain how he and his colleagues developed a lensless miniature endoscope the width of just a few human hairs that is capable of producing high-quality color images of what's going on inside animal test subjects. He likens the imaging technique to that of a pinhole camera capturing a nature scene. Hence, the tree.
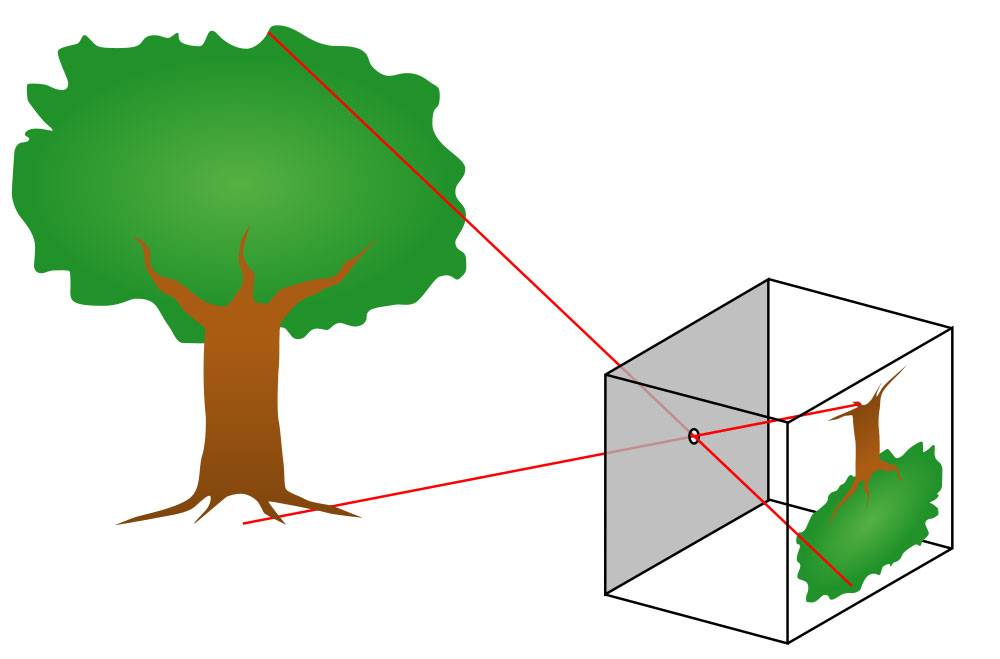
Image caption: How a pinhole camera works
Image credit: Wikimedia Commons
A pinhole camera relies not on a lens but on a tiny aperture—a screen that blocks all light except for what passes through a tiny hole. The light is scattered from each point in the scene, from the roots to the branches to the leaves, and passes through the pinhole, recreating the image, inverted. He draws a second scribble of an upside-down tree.
For their minimally invasive microendoscope, Foster and a team of Johns Hopkins engineers miniaturized that concept down to a multicore fiber just 300 microns in diameter. The novel piece of their tech is that they then used what's called a coded aperture. Instead of one pinhole, there are many randomly positioned holes on a tiny disc at one end of the fiber, each one projecting the same image, one atop another, albeit shifted ever so slightly up, down, left, right. "And so what you see is that every single one of these holes creates another copy of the image, and they're all over top of one another, and it looks ugly. It doesn't look like a tree anymore." Or, in this case, a mouse brain synapse. To the human eye, it looks like an extremely out-of-focus photograph. But think of this end result as a compressed JPEG image. Software developed by Whiting School Professor Trac Tran is able to unpack the plethora of information within and reconstruct a clear, detailed image.
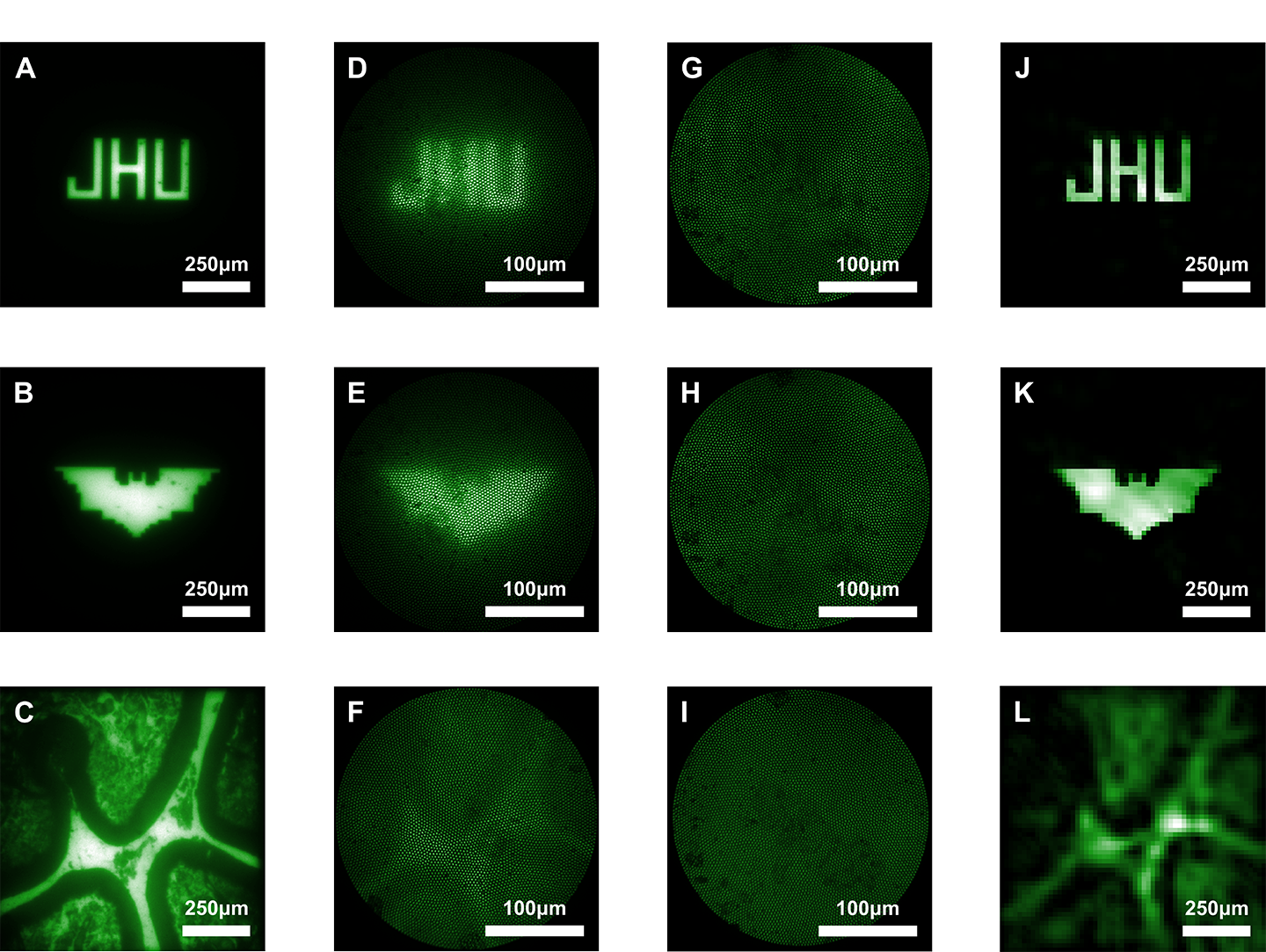
Image caption: The image above shows imaging results from the study. Images A through C show images viewed through a bulk microscope. D through F show the images as viewed through a conventional, lens-based microendoscope. G through I show the images as seen by the new lensless microendoscope. These raw images are purposefully scattered, but provide important information about light that can be used in computational reconstruction to create clearer images, shown in J through L.
Image credit: Courtesy of Mark Foster
Neuroscientists rely on microendoscopes to see inside tissue at a microscopic level—which neurons fire, for example, when a mouse is presented with a piece of cheese. Foster's new tech is a breakthrough in this type of imaging, which has traditionally offered a trade-off. Use a bulky microendoscope with a lens, and you might get a decent image, but the rigid lens damages the exact tissue you're trying to observe. That option may also inhibit movement or interfere with the subject's neural response. Use an existing lensless microendoscope and you're sacrificing image quality, especially when it's moved to focus on objects at different depths. The device developed by Foster's team produces better images than even a lens-based approach, while being thin and flexible enough to allow the subject to move freely. And it doesn't need to be moved. Their software is able to take in all the data points and make sense of where the light—an LED shone through the fiber onto neurons tagged with fluorescence—originated. "You can use a computer algorithm to reconstruct what we conventionally would see as an image, with a lot more points of information than pixels that we actually collect," he says.







