For Ashley Kiemen, it's about opening up the study of human anatomy to a whole new world.
As co-developer of CODA, a cutting-edge computational platform, she has given researchers a tool that transforms slides of human tissue into 3D maneuverable models of what happens inside organs.
"We use CODA to study human anatomy at the single cell level in ways that people hadn't been able to until now," said Kiemen, a post-doctoral fellow in the Whiting School of Engineering.
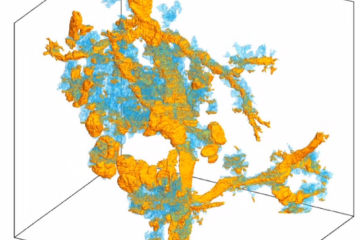
3D reconstruction of 38 PanIN (precursors to pancreatic cancer) in >2cm3 human tissue pic.twitter.com/6WMUAD0JGL
— Ashley Kiemen (@AshleyKiemen) April 2, 2021
The platform Kiemen created with Pei-Hsun Wu, an associate research professor in Johns Hopkins University's Institute for NanoBioTechnology, will be a core feature of the new Johns Hopkins Center for 3D Multiscale Cancer Imaging—a collaboration with Yale University where oncologists, pathologists, engineers, and computer scientists work together to better understand how cancer tumors form.
What distinguishes CODA from other 3D serial sectioning programs, according to Kiemen, is how it uses artificial intelligence to identify and tabulate microscopic components of human tissue.
"We use AI to label structures such as blood vessels and epithelial tissues and fat cells," she said. "By doing that, we can begin to actually quantify these things instead of just making a nice 3D movie."
Philosophizing about science
All of this is a long way from where Kiemen, a Minnesota native, began her academic career at the University of Michigan. Her first thought was to major in English, considering how much she liked those classes in high school. Then she did a Google search.
"I was looking up jobs you got if you studied English, and it was between getting a PhD and teaching or getting a business job where you might use what you learned in college, but it might not actually be that useful," she recalled.
So Kiemen shifted to STEM courses, which seemed to offer better career prospects. After she got her bachelor's degree in chemical engineering in 2016, however, she took advantage of a Michigan program—the Roger M. Jones Fellowship—through which science majors can take humanities courses at a university in European Union countries. She chose to study the philosophy of science at the London School of Economics.
Her courses there addressed what she described as "useful philosophy." She was intrigued by the distinction between deductive logic associated with scientific research and inductive logic used in day-to-day life. For her thesis, she argued that ultimately all logic is inductive, even in science.
"Let's say you have a hypothesis that all snakes have no legs," Kiemen said. "To study that deductively you would go look at a bunch of snakes, and you don't see any with legs. But that doesn't actually mean that no snakes have legs. It just means you haven't found one."
A "cool combination"
That notion of pushing beyond convention carried over to her doctoral research at JHU, where Kiemen earned her PhD in chemical and biomolecular engineering last fall. Early on, she realized that she wasn't very good at the wet lab aspect of research—"Anything I touched died," she said—but she excelled at the computational part. She also was struck by what she saw as the stunning imagery of pathology slides.
"I liked them a lot because I'm visual," she said. "When I study, I think of things visually. I'm not good at just looking at a textbook." That whetted her interest in doing something more with microscopic photos.
"I thought it was such a cool combination of math, which I love, and these beautiful images, which I love, and tying it all into biology," she said.
The final piece of what became CODA is artificial intelligence. It provides an enormous and precise level of detail about the world inside human organs, that, along with the ability to explore 3D models, will help researchers zero in on how tumors form in a complex organ like the pancreas. It's the source of one of the deadliest cancers because tumor cells spread through the bloodstream before any symptoms occur.
The hope is that CODA will enable scientists to explore the pancreas in a way that hasn't been possible, and lead to more effective diagnosis and treatment. Kiemen offered an example of what a difference CODA can make in studying the organ at a cellular level: "One thing we've studied is this lesion in the pancreas called PanIN," she said. "Just by visualizing them, we found two different phenotypes of PanIN that people didn't know existed. Because when you look at a 2D slice of tissue, you can't tell its 3D morphology. But with 3D, you could see they were different shapes. These kind of things just pop out. When we showed this to some medical pathologists, they were like, 'Oh, this is so cool.'"
Next up, Kiemen will be joining the Department of Pathology at Johns Hopkins, where she will continue to refine CODA and carry on the research into how pancreatic cancer develops.
Tagged cancer, pancreatic cancer, coda, tumors, 3d modeling